Region pages#
Clicking on any of the region buttons will take you to a page with detailed results for that region.
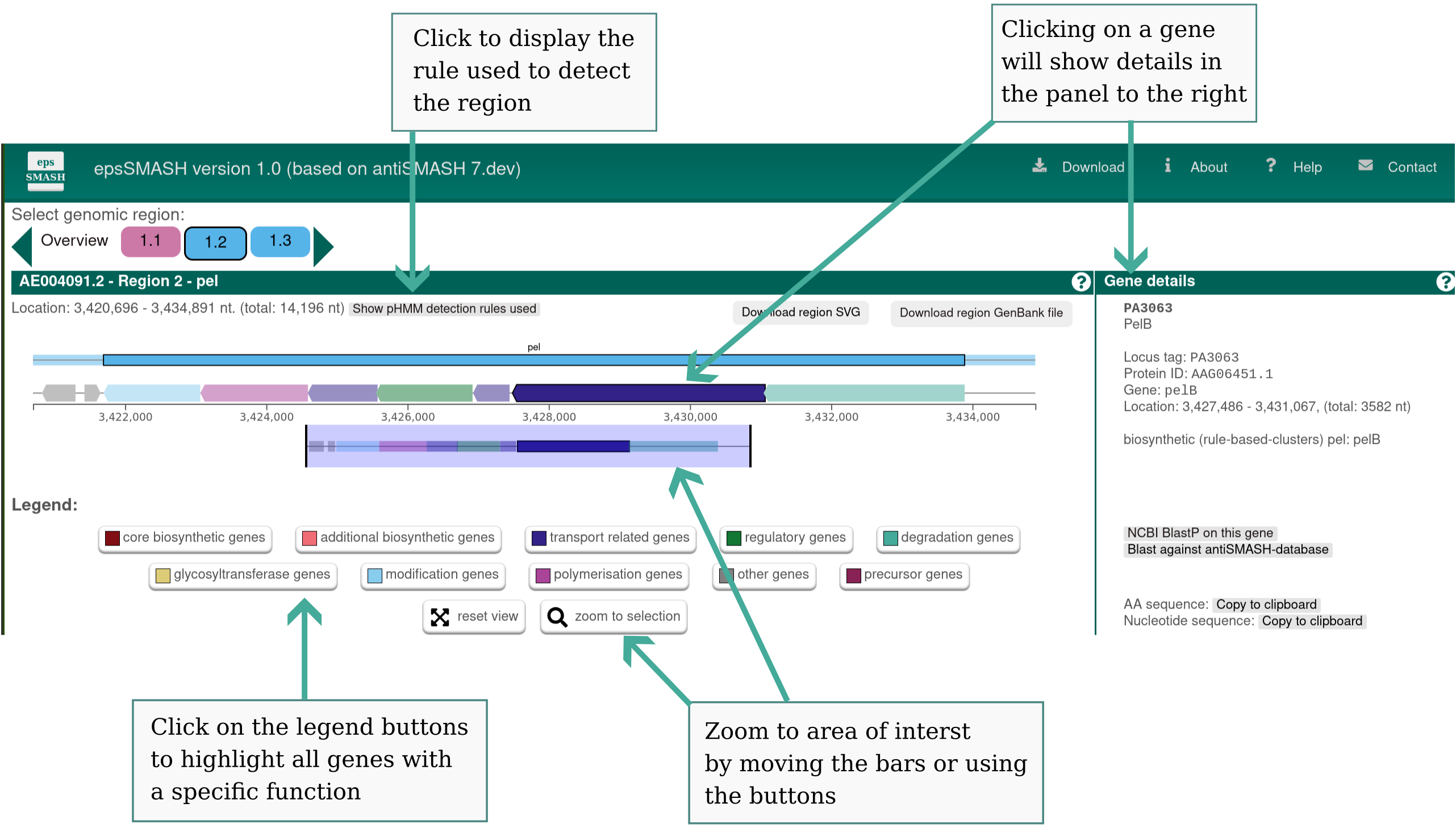
Genes are color coded by predicted function:
Blue: EPS transport (e.g. porins, scaffolding proteins)
Green: Regulation (e.g. c-di-GMP-binding proteins)
Teal: EPS degradation (e.g. lyases)
Yellow: Glycosyltransferases
Light blue: Modification (e.g. acetyltransferases)
Purple: Polymerisation (e.g. alginate synthase, Wzy)
Dark red: Precursor genes (e.g. nucleotide sugar synthesis)
If a gene does not have a known function, but is a known part of an EPS pathway, it is colored dark red. If such a gene is detected, but not part of the detected region (e.g. is detected in the borders of the region), it is colored light red. Hovering over a gene with the mouse will prompt the gene name to be displayed above the gene.
Gene details#
Clicking a gene will provide more information on the gene: identifiers, existing product annotation, gene functions, its location, and buttons for searching that gene against NCBI or the internal epsSMASH database.
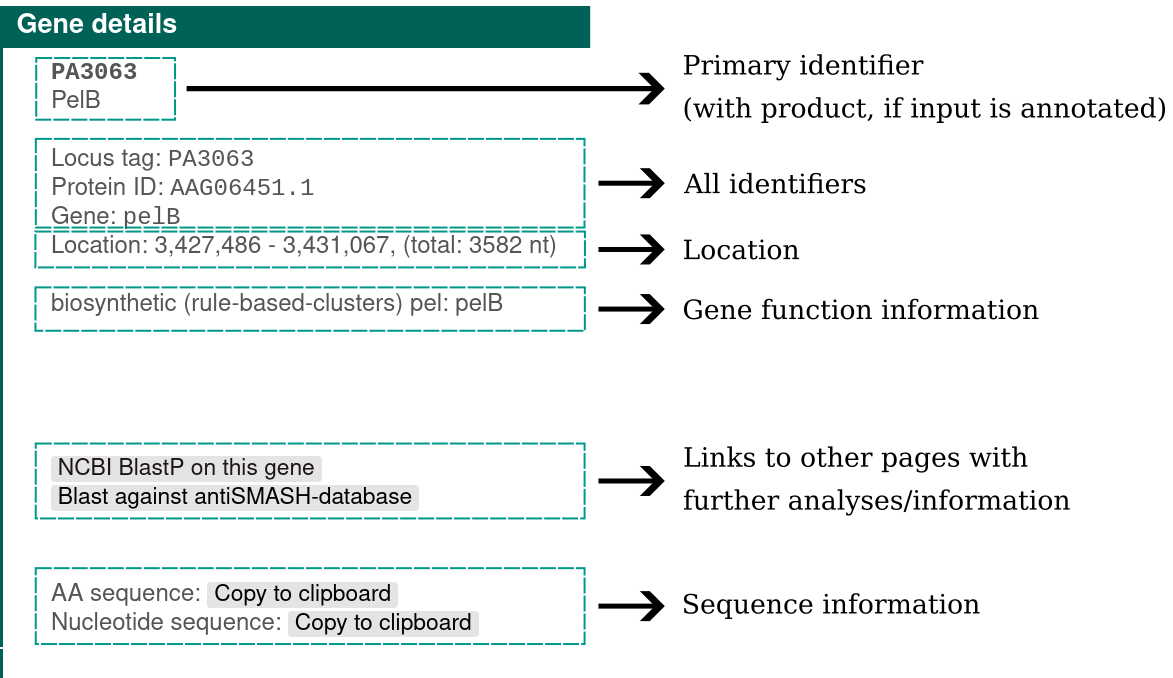
Detailed annotation#
In the bottom left panel, you can find more in-depth information on the selected gene cluster.
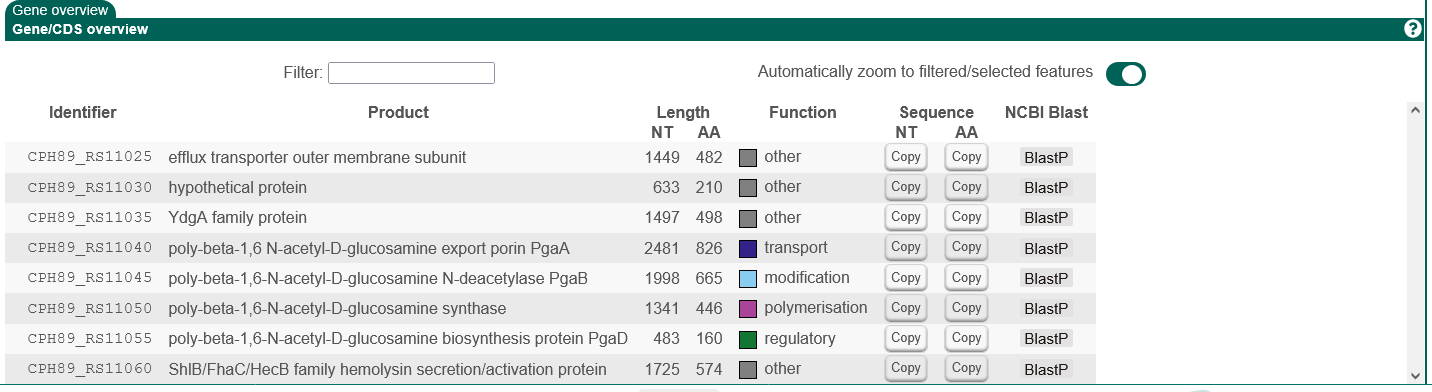
Individual prediction tools will add tabs here to provide their results.
Identifying similar BGCs#
The tab "ClusterBlast", displays the top ten gene clusters from the internal epsSMASH database that are most similar to a detected gene cluster, visually aligned to it. The drop-down selection menu can be used to browse through the gene clusters. Genes with the same colour are putative homologs based on significant Blast hits between them. Read more about ClusterBlast here.
Transcription Factor Binding Sites prediction#
The Transcription Factor Binding Sites tab is explained on its own page.
Downloading results#
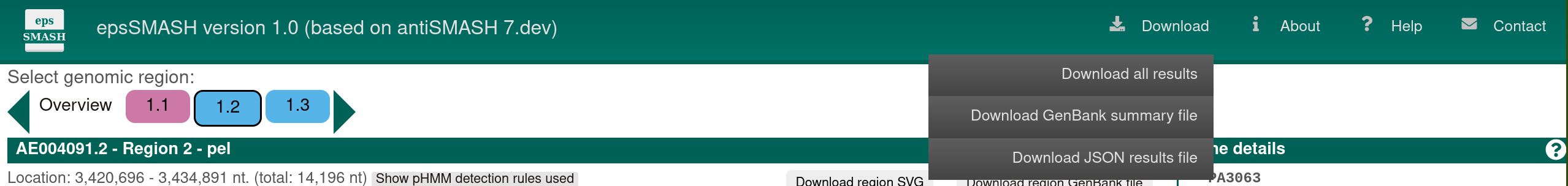
In the upper right, a small list of buttons offers further functionality.
The Help button will get you to this help page.
The About button leads to a page with information about antiSMASH, including publications.
The Download button will open a menu offering to download the complete set of results from the antiSMASH run, a summary GenBank output file, or a log file for debug purposes. The GenBank file can be viewed in a genome browser such as Artemis.
The Contact button leads to a page where you can submit support tickets to the epsSMASH developers.
Results on the public webserver are only kept for ONE month and will be deleted afterwards.
It is highly recommended that you download the full result set before they expire.